Adverse drug reactions detected on social media with natural language processing
Cancer patients undergoing chemotherapy often share word of their adverse reactions to the drugs in online health forums. Researchers at Stanford have used natural language processing to mine these posts, accurately flagging detrimental side effects well before clinical journals advise caution.
Corresponding author Julia Ransohoff, MD, and colleagues describe their work in a study published June 3 in JMIR Public Health and Surveillance.
The team drew mentions of adverse drug reactions (ADRs) from patient posts at Inspire.com related to two categories of chemo drug.
Using a signal-generation pipeline built with natural language processing, they analyzed mentions of these drugs to determine if they were causing adverse reactions in significant swaths of the relevant patient populations.
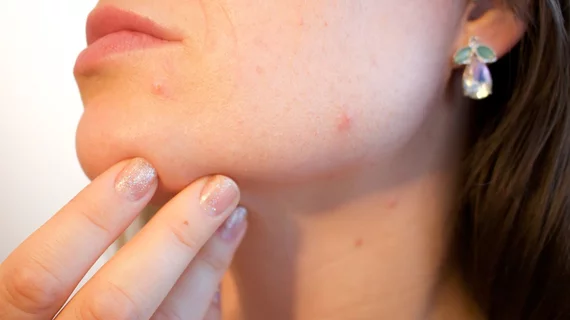
The researchers focused on skin-related reactions such as rash, acne, blisters and psoriasis.
They found their system detected these sorts of ADRs in patient reports with 90% accuracy.
Their results were later validated by mentions of the same ADRs in studies published with similar frequencies in peer-reviewed journals. And these publications were, on average, seven months behind the researchers’ findings.
The team additionally discovered one adverse reaction that affected 23 patients yet was not mentioned in 15 years of literature on the culprit drug. Ransohoff and colleagues filled the gap from their research project, reporting the finding in a clinical oncology journal.
“Several hundred million patients report health concerns in social health networks, yet this information is markedly underutilized for pharmacosurveillance,” the authors write in their discussion. “Our findings suggest the important contributions that social health network data can play in contributing to more comprehensive and timely pharmacovigilance.”
The study is available in full for free.